Phylo2021
Phylogenetic reconstruction workshop, Tartu, 2021
This project is maintained by Mycology-Microbiology-Center
0. Prerequisites
For Windows users, we suggest to use WinSCP for file transfer and PuTTY for starting a command line session on a high-performance computing (HPC) cluster.
Alternatively, on Windows 10 one may try to use WSL - Windows Subsystem for Linux (see installation guide here).
For Linux and Mac users, it would be possible to use a terminal emulator.
We’ll run analysis on the Rocket Cluster of the University of Tartu.
Windows users, WinSCP setup
-
To connect to the server, add your user and password as shown on the image:
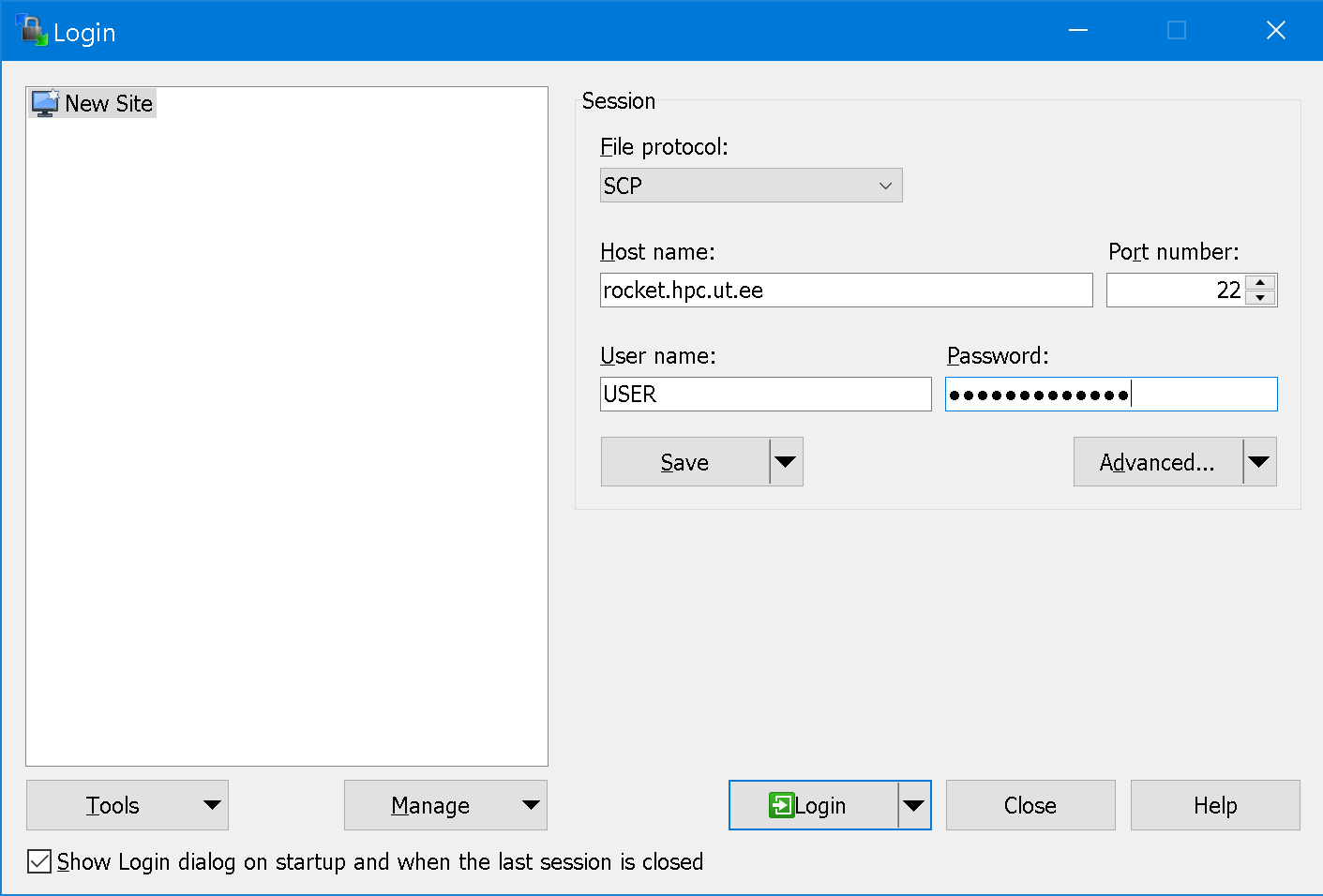
File protocol: SCP
Host name:rocket.hpc.ut.ee
User name: substituteUSERwith your login -
To setup automatic transmission of WinSCP passwords to PuTTY (SSH), go to Menu
Options->Preferences->ApplicationsSet the tickmark onRemeber session password and pass it to PuTTY (SSH)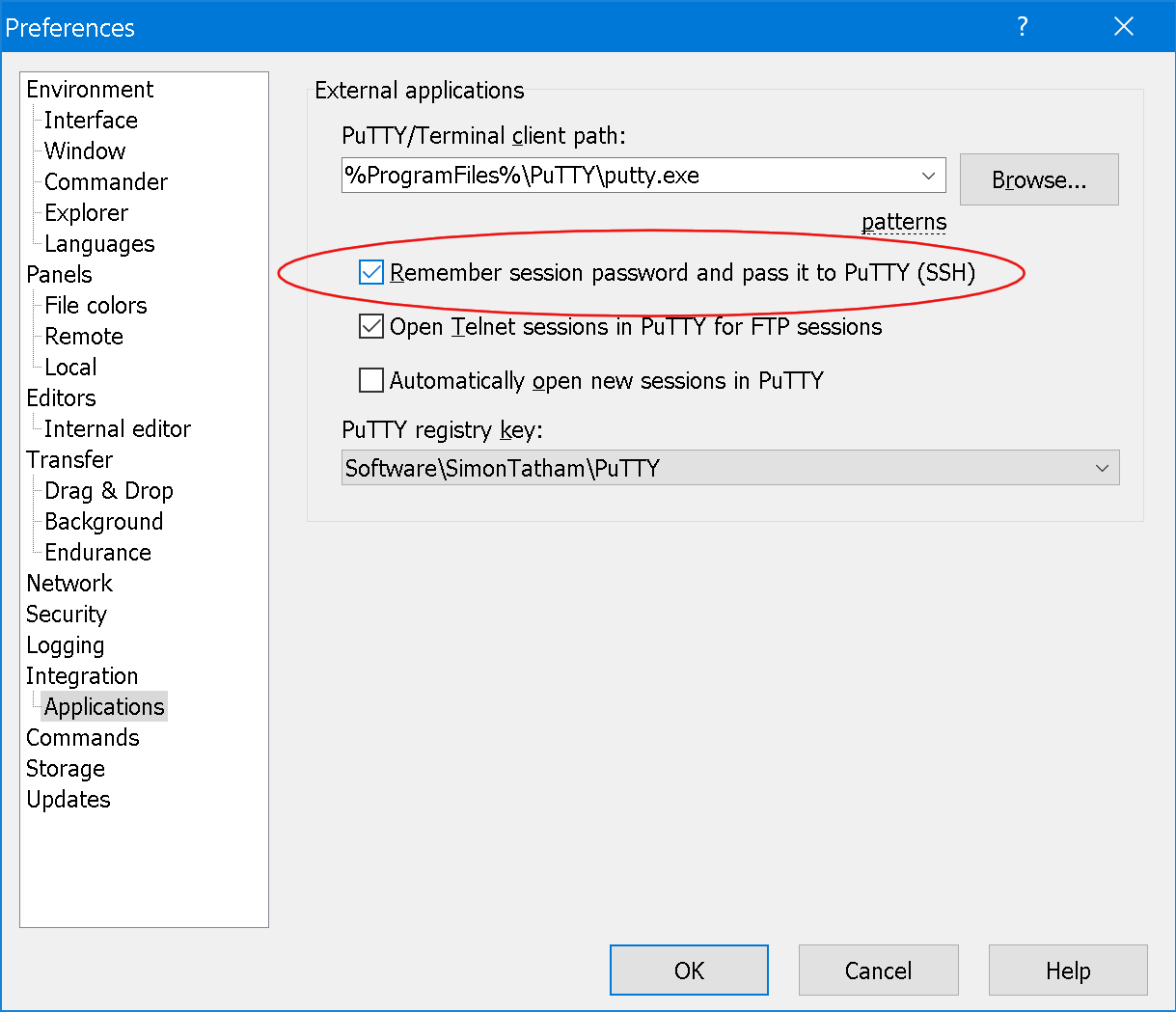
-
To start a command line session on HPC cluster, press
Ctrl + Por go to menuCommands->Open in PuTTY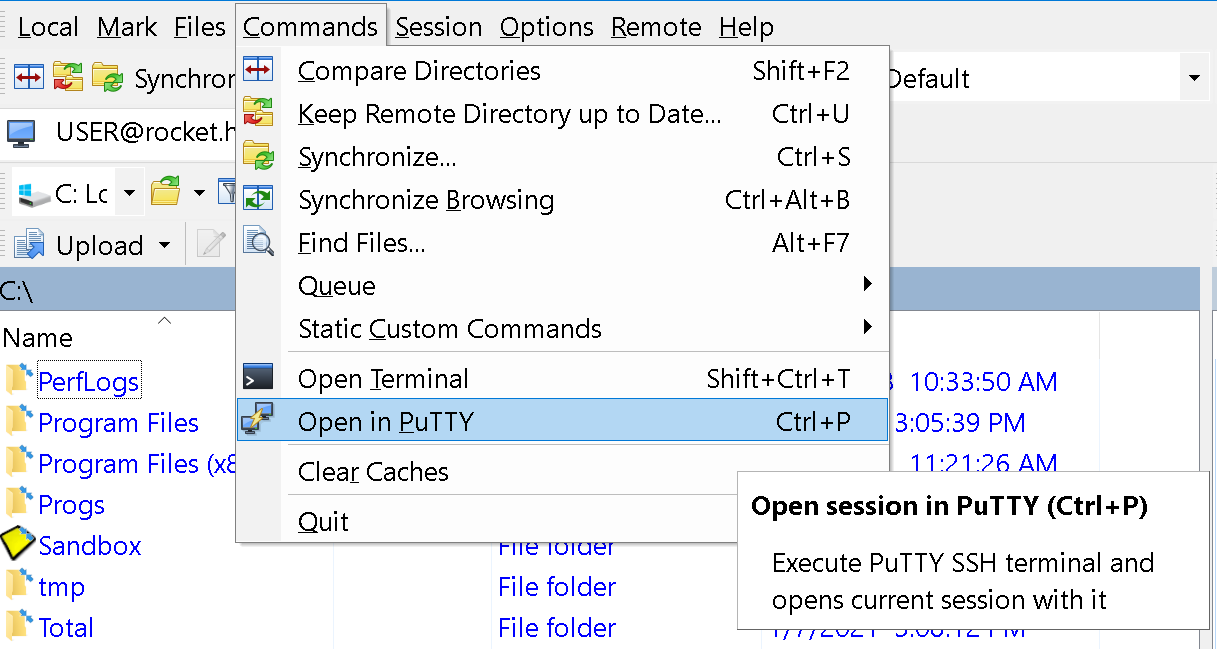
For Linux and Mac users
To connect to to the HPC cluster just run:
ssh USER@rocket.hpc.ut.ee
(substitute USER with your login).
To copy a file or multiple files to HPC cluster use:
scp yourfile amiri@rocket.hpc.ut.ee:~/yourfile # single file
scp file1 file2 amiri@rocket.hpc.ut.ee:~/ # multiple files
To copy file from HPC cluster (e.g., yourfile from home directory on HPC to home directory on your computer) use:
scp USER@rocket.hpc.ut.ee:~/yourfile ~/yourfile
If you have large files (or large number of files), it’s better to use rsync program for file transfer, e.g.
rsync -avz Documents/* user@rocket.hpc.ut.ee:~/all/
Logout from HPC cluster
When you are done, to end your session on HPC cluster run:
exit
1. Setup working environment on HPC cluster
Conda
On the HPC systems and clusters, users can install software into their home directory where they have write permissions. To make the life easier, you may use Conda - a package manager, which helps you find and install the software and its dependencies.
To install Miniconda run the following code:
wget https://repo.anaconda.com/miniconda/Miniconda3-latest-Linux-x86_64.sh -O ~/miniconda.sh
bash ~/miniconda.sh -b -p $HOME/miniconda
~/miniconda/bin/conda init bash
source ~/.bashrc
conda update --all --yes -c bioconda -c conda-forge
conda install --yes -c conda-forge mamba
For phylogenetic tree inference, we will use the following programs on the cluster:
To install the software run*:
conda install -c bioconda -c conda-forge raxml-ng mrbayes
* Probably this step will not work on Rocket Cluster due to the conflict of versions. To overcome this issue, we will create a separate environment with conda.
Conda environments
If the software you whish to use could not be installed to the base (default) environment due to the conflict of versions, or you want to use a specific version of the program, or just want to keep it separate, you may create a separate envrionment with:
mamba create --name PHYLO -c bioconda -c conda-forge mrbayes=3.2.6 raxml-ng
conda activate PHYLO # swith to the new environment we've created
Verify which software version are installed:
raxml-ng --version
mb about
To swith to the base environment run:
conda deactivate
Module system
Alternatively, if the software you whish to use is pre-installed on the HPC cluster, you may load it as an environment module.
To list all available modules, use module avail command (scroll the list with space button, press q to quit).
To search for a particular module, use e.g. module -r spider '.*mrbayes.*'.
If the required software was found, you need to load the module, e.g.:
module load mrbayes/3.2.7a
mb about # verify if MrBayes is available
If you’ve set up the access to the HPC cluster and installed the software you need, you may proceed to the next part:
2. Scheduling jobs on HPC