Phylo2021
Phylogenetic reconstruction workshop, Tartu, 2021
This project is maintained by Mycology-Microbiology-Center
4. Visualization of phylogenetic trees: preparation
This is also available as an R Script file: scripts_tree_operations.R
A. Setting up an RStudio project
For convenience, we suggest working with R from RStudio. Before starting, make a directory for your project (e.g. phylo_workshop), create folder data inside of it, and put two trees and tables for label renaming in data. Then, in R-Studio go to File -> New Project -> Existing Directory and select phylo_workshop you’ve just created. To make a place for your code, go File -> New File -> R Script and save it in the project directory.
B. Importing trees to the R environment
To handle trees we will primarily use popular packages treeio (for tree import and format conversions) and ggtree (for visualizations). For documentation on these, see https://yulab-smu.top/treedata-book/index.html. ggtree produces plots using ggplot2 capabilities and syntax. We also will use a few functions from ape and geiger - two other widely used phylogenetic packages. If you do not have them installed, it can be done as follows:
install.packages(c('ape', 'ggplot2', 'geiger'))
## ggtree and treeio come through Bioconductor:
if (!requireNamespace("BiocManager", quietly = TRUE))
install.packages("BiocManager")
BiocManager::install(c('treeio', 'ggtree'))
Load these packages into your environment:
library(treeio)
library(ggtree)
library(ape)
library(ggplot2)
library(geiger)
Tree import
Read maximum likelihood tree with bootstraps (RAxML-NG) and Bayesian tree with posterior probabilities (MrBayes). Later we will use Bayesian tree as a basis of visualization, and RAxML-NG tree to obtain bootstrap values.
bootTree <- read.newick ('data/tree_raxml.nwk')
bayesTree <- read.nexus ('data/tree_mrbayes.nex')
Bayesian tree file includes 2 trees: first with posteriors, and second without them (topology only). We will operate on the first.
bayesTree <- bayesTree[[1]]
Visual check of the trees
Here you can see a typical ggplot2-styled syntax defining parts of the graph. ggtree itself draws a naked tree, geom_tiplab adds labels to tips, geom_text2 adds text to the plot - in this case support values stored in label field, and xlim is needed to shrink tree horizontally, to avoid truncation of long labels.
ggtree(bootTree) +
geom_tiplab() +
geom_text2(
aes(label = label, subset = !isTip),
hjust = 1.3,
vjust = -0.4,
size = 2
) +
xlim(0, 0.3)
ggtree(bayesTree) +
geom_tiplab() +
geom_text2(
aes(label = round(as.numeric(label), 2), subset = !isTip),
hjust = 1.3,
vjust = -0.4,
size = 2
) +
xlim(0, 0.3)
C. Detour: how to deal with default formatting of MrBayes files
TL;DR: Default MrBayes/BEAST output needs special transformations in R for downstream plotting. If you brought your own trees made before workshop, you likely need to read this.
Newick and NEXUS are the most common tree formats, and in both of them any internal-node-associated data (like bootstraps) can be stored as node names. In this case, it’s safe to read a tree in a simple phylo format. Though, many Bayesian programs produce a complex output that cannot fit in the node names, and instead they write it in node comments. Here, using phylo may result in a loss of comments (and node.label in phylo will be absent). So it’s better to employ more sophisticated formats: e.g. treedata used by treeio/ggtree. To make sure comments are not lost, do not use generic read.nexus, but try to find a function designed specifically for your program (see https://yulab-smu.top/treedata-book/chapter1.html).
MrBayes by default saves trees in a format friendly for FigTree (conformat=Figtree in MrBayes block), where posteriors are stored in comments (see square brackets in NEXUS). For this workshop we are not interested in FigTree compatibility, so we used comments-free conformat=simple. Though, your older trees may have been formatted for FigTree, and to read these properly, execute:
bayesTree <- read.mrbayes ('data/tree_mrbayes_figtree.nex')
This will produce a treedata object, that consists mainly of 2 parts: familiar phylo part with tree topology, and data with MrBayes-specific data including posteriors. Importantly, our plotting routine requires posteriors inside of phylo, and we achieve this with the following. (Below is MrBayes version, but with few adjustments it also works for BEAST)
## Sort nodes in data to match node order in phylo.
bayesTree@data <-
bayesTree@data[order(as.numeric(bayesTree@data$node)), ]
## Write posteriors as node labels. If tree is already rooted, do not append '' to vector.
bayesTree@phylo$node.label <-
append ('', as.vector(subset(
bayesTree@data$prob,
as.numeric(bayesTree@data$node) > length(bayesTree@phylo$tip.label)
)))
## Translate treedata into phylo.
bayesTree <- as.phylo(bayesTree)
Alternatively, you can convert your trees from nexus to newick without loosing probabilites with a free software TreeGraph. See the tutorial video.
D. Rerooting
A common practice is to reroot a tree on outgroup. For the next step we need to root both trees. To do so, you need to know either:
- Labels of outgroup taxa, or
- An ID of a node joining all the taxa belonging to the outgroup.
1. Rerooting by specifying outgroup labels
## Bayesian tree:
bayesTree_rooted <-
ape::root(bayesTree,
outgroup = c('MM35985', 'MYX328'),
edgelabel = TRUE)
## Same for ML tree:
bootTree_rooted <-
ape::root(bootTree,
outgroup = c('MM35985', 'MYX328'),
edgelabel = TRUE)
Note: edgelabel = T is essential for correct label placement, otherwise labels can slip to wrong branches. Sometimes you can spot slippage by ingroup and outgroup that have different support values after rerooting, and by missing values on branches:
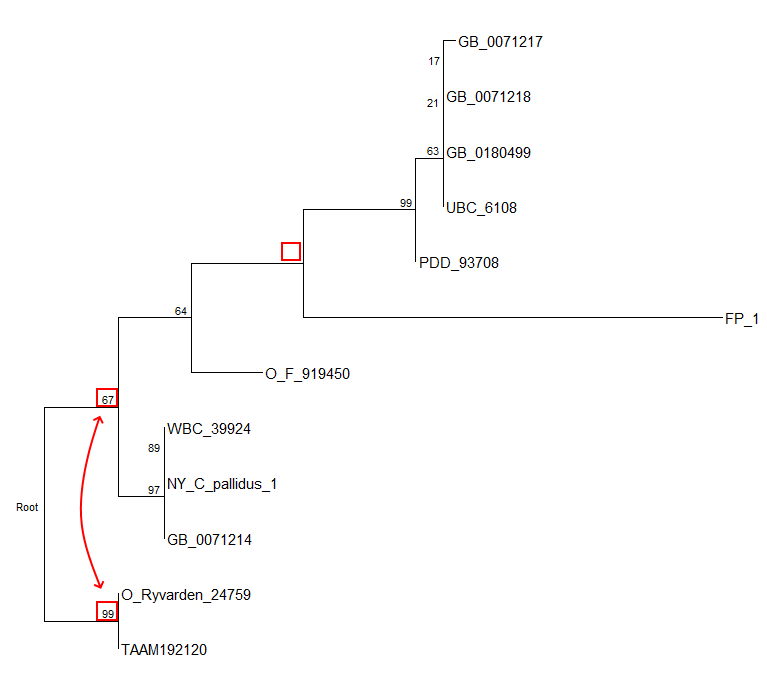
For details consult Czech et al. 2017 “A Critical Review on the Use of Support Values in Tree Viewers and Bioinformatics Toolkits”.
2. Rerooting by specifying node ID
If outgroup is large, the option with node ID may save you some typing. The easiest way to locate needed ID is to plot all of them (here on example of the Bayesian tree).
ggtree(bayesTree) +
geom_tiplab() +
geom_text(
aes(label = node),
hjust = 1.3,
vjust = 1.3,
size = 2,
color = 'red'
) +
xlim(0, 0.3)
If your tree is large and crowded, it may be difficult to visually spot the desired node. Another way is to make a table from the tree, find a taxon from the outgroup and trace node-to-node connections down to the common node of all outgroup taxa.
bayesTree_tib <- as_tibble(bayesTree)
In this case it’s node 89:
bayesTree_rooted <- ape::root(bayesTree, node = 89, edgelabel = TRUE)
Check the result
ggtree(bootTree_rooted) +
geom_tiplab() +
geom_text2(
aes(label = round(as.numeric(label), 2), subset = !isTip),
hjust = 1.3,
vjust = -0.4,
size = 2
) +
xlim(0, 0.3)
ggtree(bayesTree_rooted) +
geom_tiplab() +
geom_text2(
aes(label = round(as.numeric(label), 2), subset = !isTip),
hjust = 1.3,
vjust = -0.4,
size = 2
) +
xlim(0, 0.3)
E. Combine bootstraps and posteriors
Set up a function
The getSupports function compares all the subclades in the Bayes tree to all the subclades in the bootstrap tree, and vice versa, and identifies all those clades that are identical. It produces a data frame supportsTable with node IDs and their respective bootstraps for further use in ggtree.
The function originates from Ordynets A., 2018 with slight changes by Yatsiuk I.
Let’s fetch it from GitHub:
source('https://github.com/Mycology-Microbiology-Center/Phylo2021/blob/main/scripts/getSupports.R?raw=TRUE')
getSupports should appear in your environment. Then apply it to the trees, where primaryTree is what we want to plot in the end:
supportsTable <- getSupports(primaryTree = bayesTree_rooted, secondaryTree = bootTree_rooted)
Next step is: 5. Visualization of phylogenetic trees: annotation.